32. AR(1) Processes
32.1Overview¶
In this lecture we are going to study a very simple class of stochastic models called AR(1) processes.
These simple models are used again and again in economic research to represent the dynamics of series such as
- labor income
- dividends
- productivity, etc.
We are going to study AR(1) processes partly because they are useful and partly because they help us understand important concepts.
Let’s start with some imports:
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (11, 5) #set default figure size32.2The AR(1) model¶
The AR(1) model (autoregressive model of order 1) takes the form
where are scalar-valued parameters
(Equation (1) is sometimes called a stochastic difference equation.)
The specification (1) generates a time series as soon as we specify an initial condition .
To make things even simpler, we will assume that
- the process is IID and standard normal,
- the initial condition is drawn from the normal distribution and
- the initial condition is independent of .
32.2.1Moving average representation¶
Iterating backwards from time , we obtain
If we work all the way back to time zero, we get
Equation (3) shows that is a well defined random variable, the value of which depends on
- the parameters,
- the initial condition and
- the shocks from time to the present.
Throughout, the symbol will be used to refer to the density of this random variable .
32.2.2Distribution dynamics¶
One of the nice things about this model is that it’s so easy to trace out the sequence of distributions corresponding to the time series .
To see this, we first note that is normally distributed for each .
This is immediate from (3), since linear combinations of independent normal random variables are normal.
Given that is normally distributed, we will know the full distribution if we can pin down its first two moments.
Let and denote the mean and variance of respectively.
We can pin down these values from (3) or we can use the following recursive expressions:
These expressions are obtained from (1) by taking, respectively, the expectation and variance of both sides of the equality.
In calculating the second expression, we are using the fact that and are independent.
(This follows from our assumptions and (3).)
Given the dynamics in (3) and initial conditions , we obtain and hence
The following code uses these facts to track the sequence of marginal distributions .
The parameters are
a, b, c = 0.9, 0.1, 0.5
mu, v = -3.0, 0.6 # initial conditions mu_0, v_0Here’s the sequence of distributions:
from scipy.stats import norm
sim_length = 10
grid = np.linspace(-5, 7, 120)
fig, ax = plt.subplots()
for t in range(sim_length):
mu = a * mu + b
v = a**2 * v + c**2
ax.plot(grid, norm.pdf(grid, loc=mu, scale=np.sqrt(v)),
label=fr"$\psi_{t}$",
alpha=0.7)
ax.legend(bbox_to_anchor=[1.05,1],loc=2,borderaxespad=1)
plt.show()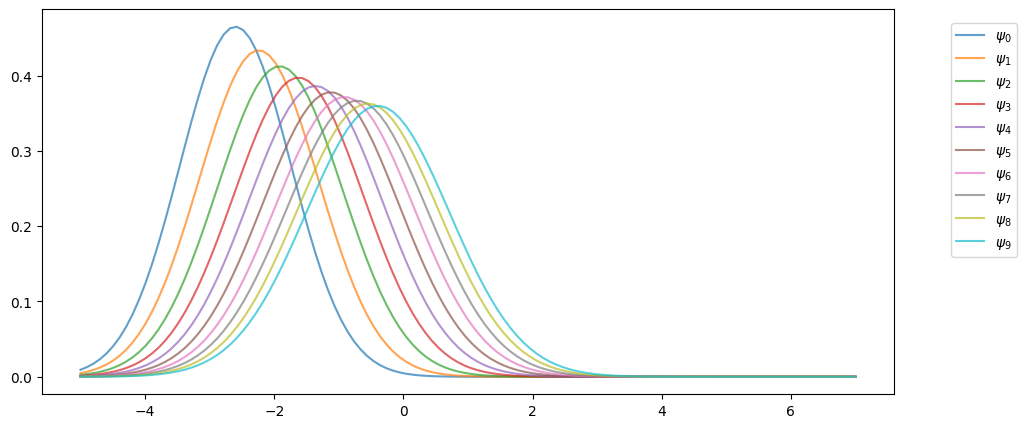
32.3Stationarity and asymptotic stability¶
When we use models to study the real world, it is generally preferable that our models have clear, sharp predictions.
For dynamic problems, sharp predictions are related to stability.
For example, if a dynamic model predicts that inflation always converges to some kind of steady state, then the model gives a sharp prediction.
(The prediction might be wrong, but even this is helpful, because we can judge the quality of the model.)
Notice that, in the figure above, the sequence seems to be converging to a limiting distribution, suggesting some kind of stability.
This is even clearer if we project forward further into the future:
def plot_density_seq(ax, mu_0=-3.0, v_0=0.6, sim_length=40):
mu, v = mu_0, v_0
for t in range(sim_length):
mu = a * mu + b
v = a**2 * v + c**2
ax.plot(grid,
norm.pdf(grid, loc=mu, scale=np.sqrt(v)),
alpha=0.5)
fig, ax = plt.subplots()
plot_density_seq(ax)
plt.show()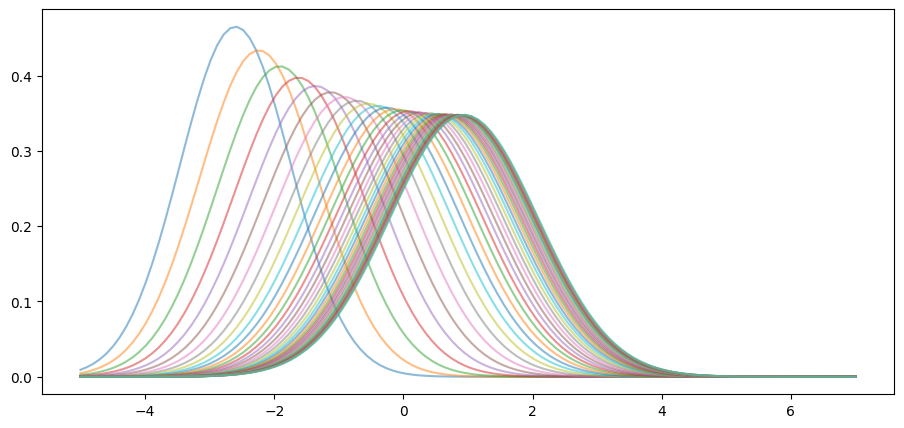
Moreover, the limit does not depend on the initial condition.
For example, this alternative density sequence also converges to the same limit.
fig, ax = plt.subplots()
plot_density_seq(ax, mu_0=4.0)
plt.show()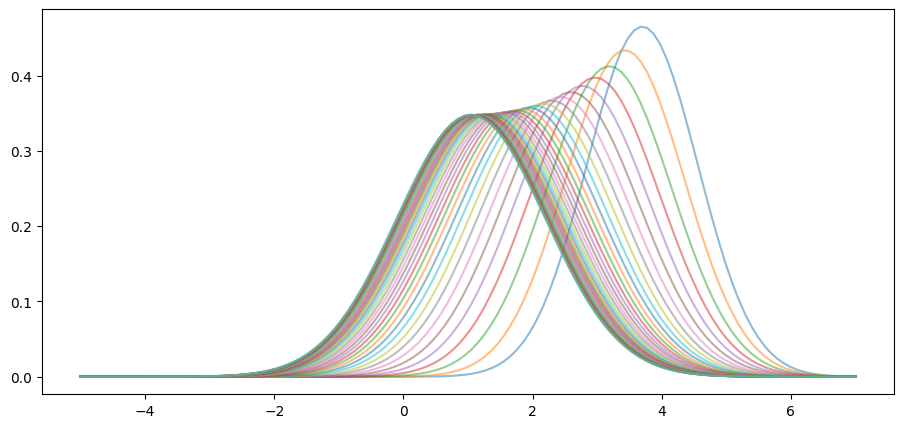
In fact it’s easy to show that such convergence will occur, regardless of the initial condition, whenever .
To see this, we just have to look at the dynamics of the first two moments, as given in (4).
When , these sequences converge to the respective limits
(See our lecture on one dimensional dynamics for background on deterministic convergence.)
Hence
We can confirm this is valid for the sequence above using the following code.
fig, ax = plt.subplots()
plot_density_seq(ax, mu_0=4.0)
mu_star = b / (1 - a)
std_star = np.sqrt(c**2 / (1 - a**2)) # square root of v_star
psi_star = norm.pdf(grid, loc=mu_star, scale=std_star)
ax.plot(grid, psi_star, 'k-', lw=2, label=r"$\psi^*$")
ax.legend()
plt.show()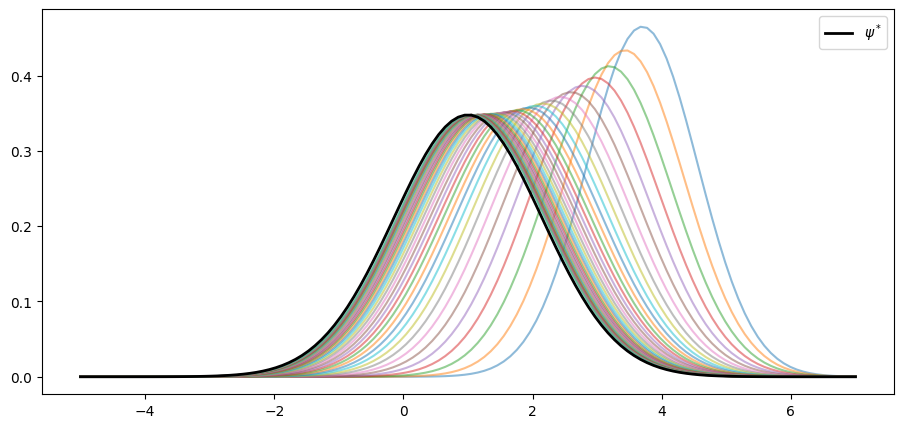
As claimed, the sequence converges to .
We see that, at least for these parameters, the AR(1) model has strong stability properties.
32.3.1Stationary distributions¶
Let’s try to better understand the limiting distribution .
A stationary distribution is a distribution that is a “fixed point” of the update rule for the AR(1) process.
In other words, if is stationary, then for all in .
A different way to put this, specialized to the current setting, is as follows: a density ψ on is stationary for the AR(1) process if
The distribution in (7) has this property --- checking this is an exercise.
(Of course, we are assuming that so that is well defined.)
In fact, it can be shown that no other distribution on has this property.
Thus, when , the AR(1) model has exactly one stationary density and that density is given by .
32.4Ergodicity¶
The concept of ergodicity is used in different ways by different authors.
One way to understand it in the present setting is that a version of the law of large numbers is valid for , even though it is not IID.
In particular, averages over time series converge to expectations under the stationary distribution.
Indeed, it can be proved that, whenever , we have
whenever the integral on the right hand side is finite and well defined.
Notes:
- In (9), convergence holds with probability one.
- The textbook by Meyn & Tweedie (2009) is a classic reference on ergodicity.
Ergodicity is important for a range of reasons.
For example, (9) can be used to test theory.
In this equation, we can use observed data to evaluate the left hand side of (9).
And we can use a theoretical AR(1) model to calculate the right hand side.
If is not close to , even for many observations, then our theory seems to be incorrect and we will need to revise it.
32.5Exercises¶
Solution to Exercise 1
Here is one solution:
from numba import njit
from scipy.special import factorial2
@njit
def sample_moments_ar1(k, m=100_000, mu_0=0.0, sigma_0=1.0, seed=1234):
np.random.seed(seed)
sample_sum = 0.0
x = mu_0 + sigma_0 * np.random.randn()
for t in range(m):
sample_sum += (x - mu_star)**k
x = a * x + b + c * np.random.randn()
return sample_sum / m
def true_moments_ar1(k):
if k % 2 == 0:
return std_star**k * factorial2(k - 1)
else:
return 0
k_vals = np.arange(6) + 1
sample_moments = np.empty_like(k_vals)
true_moments = np.empty_like(k_vals)
for k_idx, k in enumerate(k_vals):
sample_moments[k_idx] = sample_moments_ar1(k)
true_moments[k_idx] = true_moments_ar1(k)
fig, ax = plt.subplots()
ax.plot(k_vals, true_moments, label="true moments")
ax.plot(k_vals, sample_moments, label="sample moments")
ax.legend()
plt.show()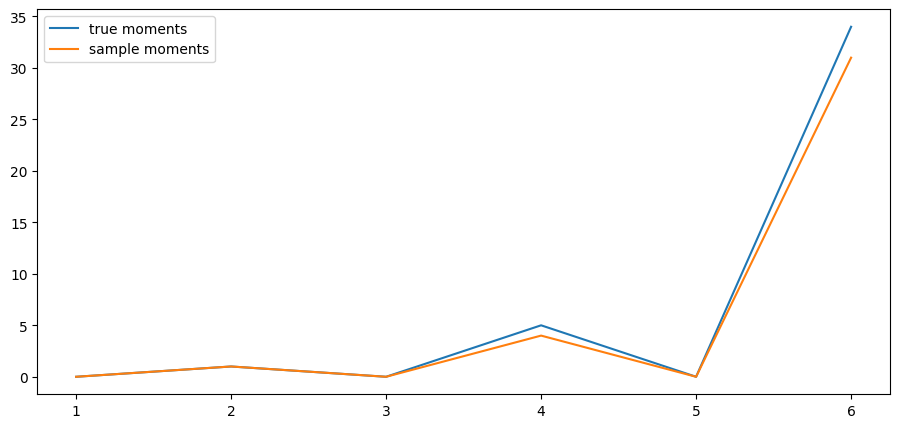
Solution to Exercise 2
Here is one solution:
K = norm.pdf
class KDE:
def __init__(self, x_data, h=None):
if h is None:
c = x_data.std()
n = len(x_data)
h = 1.06 * c * n**(-1/5)
self.h = h
self.x_data = x_data
def f(self, x):
if np.isscalar(x):
return K((x - self.x_data) / self.h).mean() * (1/self.h)
else:
y = np.empty_like(x)
for i, x_val in enumerate(x):
y[i] = K((x_val - self.x_data) / self.h).mean() * (1/self.h)
return ydef plot_kde(ϕ, x_min=-0.2, x_max=1.2):
x_data = ϕ.rvs(n)
kde = KDE(x_data)
x_grid = np.linspace(-0.2, 1.2, 100)
fig, ax = plt.subplots()
ax.plot(x_grid, kde.f(x_grid), label="estimate")
ax.plot(x_grid, ϕ.pdf(x_grid), label="true density")
ax.legend()
plt.show()from scipy.stats import beta
n = 500
parameter_pairs= (2, 2), (2, 5), (0.5, 0.5)
for α, β in parameter_pairs:
plot_kde(beta(α, β))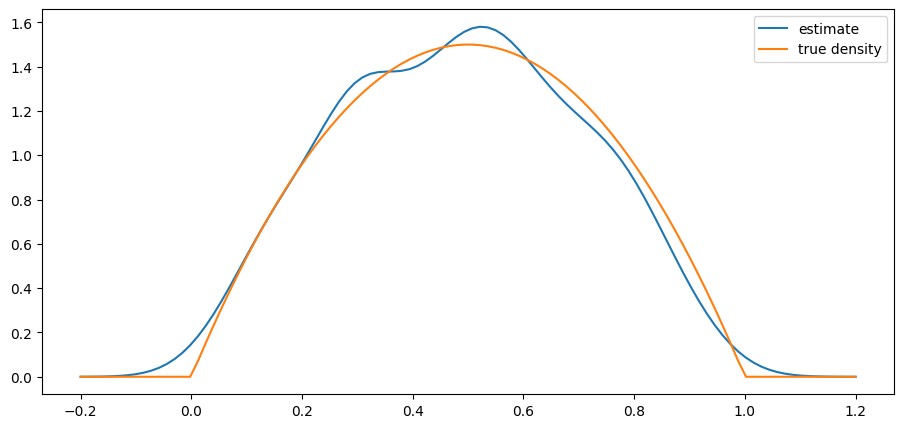
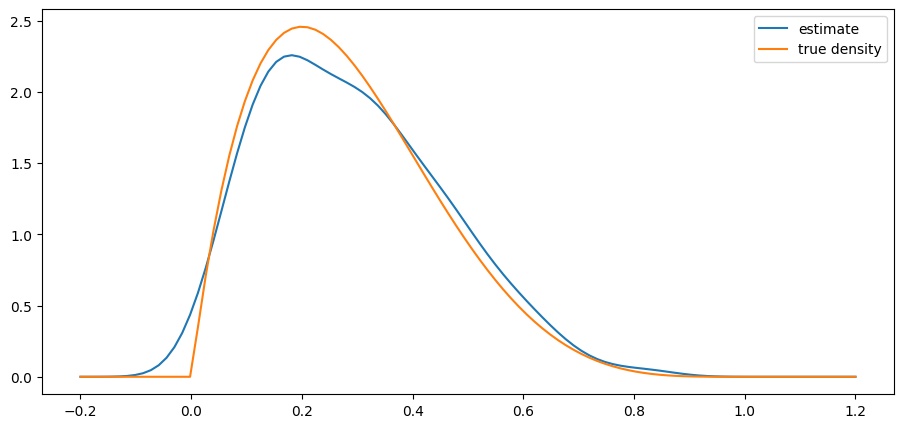
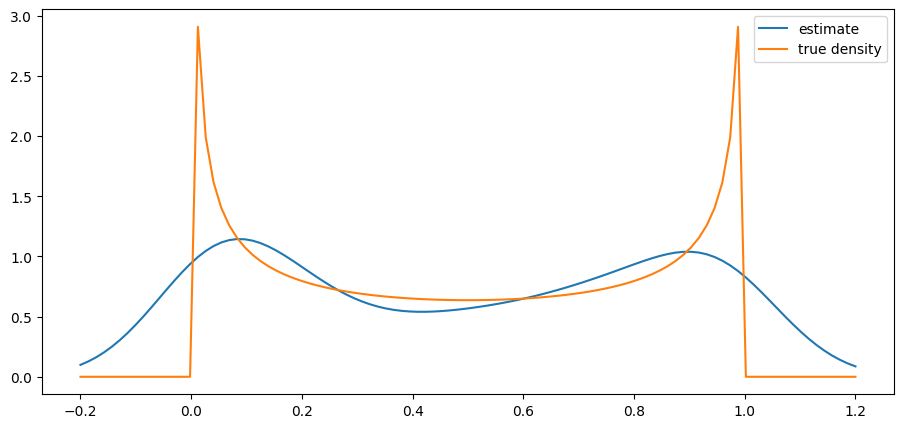
We see that the kernel density estimator is effective when the underlying distribution is smooth but less so otherwise.
Solution to Exercise 3
Here is our solution
a = 0.9
b = 0.0
c = 0.1
μ = -3
s = 0.2μ_next = a * μ + b
s_next = np.sqrt(a**2 * s**2 + c**2)ψ = lambda x: K((x - μ) / s)
ψ_next = lambda x: K((x - μ_next) / s_next)ψ = norm(μ, s)
ψ_next = norm(μ_next, s_next)n = 2000
x_draws = ψ.rvs(n)
x_draws_next = a * x_draws + b + c * np.random.randn(n)
kde = KDE(x_draws_next)
x_grid = np.linspace(μ - 1, μ + 1, 100)
fig, ax = plt.subplots()
ax.plot(x_grid, ψ.pdf(x_grid), label="$\psi_t$")
ax.plot(x_grid, ψ_next.pdf(x_grid), label="$\psi_{t+1}$")
ax.plot(x_grid, kde.f(x_grid), label="estimate of $\psi_{t+1}$")
ax.legend()
plt.show()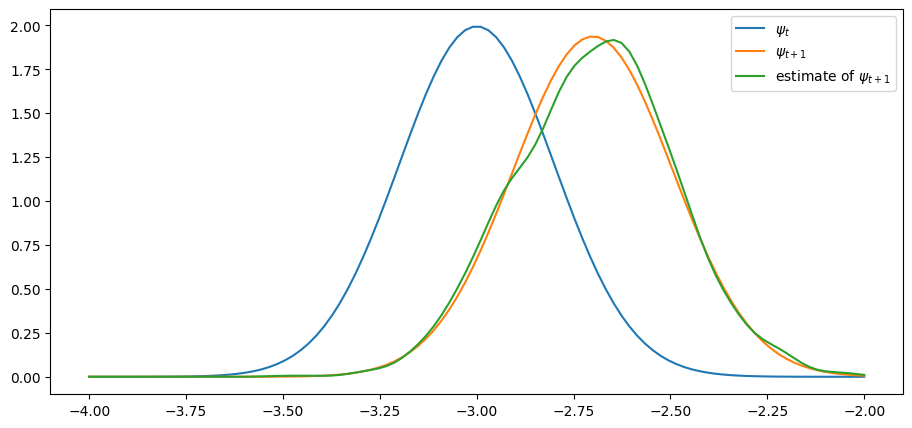
The simulated distribution approximately coincides with the theoretical distribution, as predicted.
- Meyn, S. P., & Tweedie, R. L. (2009). Markov Chains and Stochastic Stability. Cambridge University Press.

Creative Commons License – This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International.
